Project #2: CellPyAbility
CellPyAbility GitHub
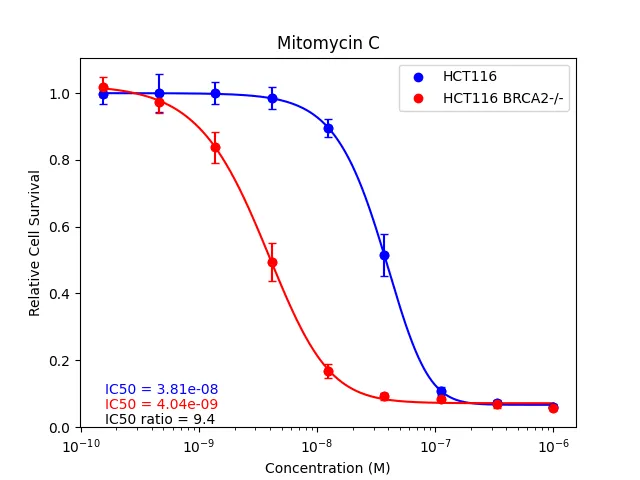
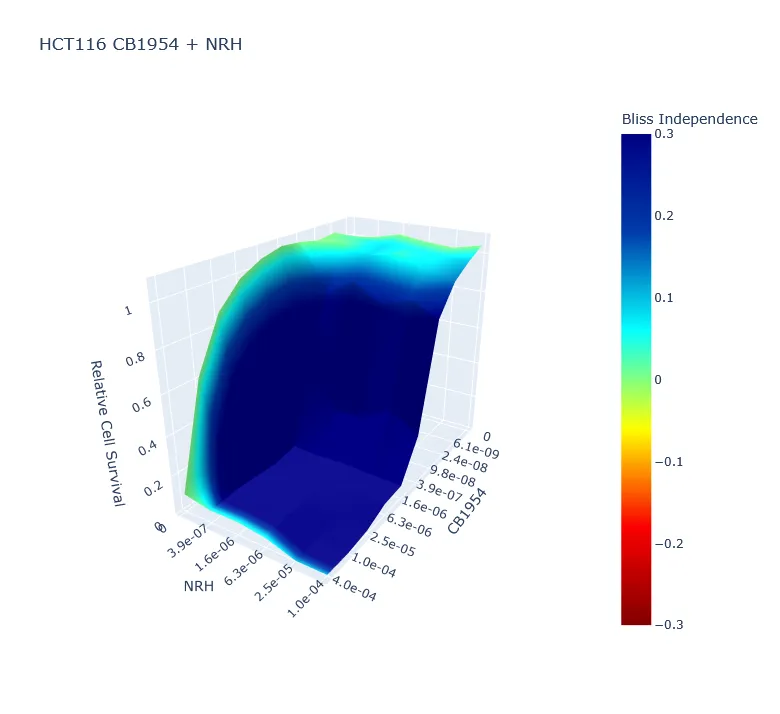
Dose-response assays measure viability across drug gradients, a fundamental tool in cell biology and drug discovery. Compared to common methods of measuring cell viability, nuclei counting provides several advantages. However, a major disadvantage of nuclei counting is the computational and temporal cost of the required image and data analyses. Accordingly, I designed CellPyAbility, a software suite that automates image processing, dose-response fitting, and synergy calculation for cell viability assays. Here's a Yale Cancer Center news article featuring our work.
CellPyAbility combines the advantages of nuclei counting with high-throughput protocols and automated analysis to substantially lower the barriers to dose-response assays. The software can be run as a code-free Windows application or platform-independent Python package. Please see CellPyAbility v0.1.0 on PyPI for package details.
This project revealed my love of programming, and I am hopelessly hooked. CellPyAbility started as a Python script I wrote to automate unwieldy data wrangling with cell viability assays, a staple of my PhD training in the Bindra Lab at Yale. As my first real programming experience, it teemed with absolute paths and repetitive code, lacked logging and proactive error handling, and generally sucked.
But it seemed good enough to win the interest of my labmates. I started writing new versions with other users in mind; this is a completely different experience from writing code for yourself. It forced me to prepare for edge cases, anticipate quality of life demands, create GUIs, be clear with my language, and continuously expand the circle of potential users. As a bench scientist of several years, engaging in a creative and user-oriented project was a delight.